Accessing Pangeo Climate Data
Accessing Pangeo Climate Data¶
In this notebook, we select and pull CESM data from Pangeo using our pip-installable library, cesm.
# Import libraries
import os
import pandas as pd
## After pip install,
from cesm import cesm_pull as cpull
# Read in available datasets, filtered for NCAR's CESM
ncar_models = cpull.find_available_data()
ncar_models.drop(columns=['zstore']) # dropping link for cleaner ouput
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|
| 198051 | CMIP | NCAR | CESM2 | historical | r11i1p1f1 | Amon | hurs | gn | NaN | 20190514 |
| 198054 | CMIP | NCAR | CESM2 | historical | r11i1p1f1 | Amon | huss | gn | NaN | 20190514 |
| 198057 | CMIP | NCAR | CESM2 | historical | r11i1p1f1 | Amon | pr | gn | NaN | 20190514 |
| 198172 | CMIP | NCAR | CESM2 | historical | r11i1p1f1 | Amon | tas | gn | NaN | 20190514 |
| 437199 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | huss | gn | NaN | 20200528 |
| 437549 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | tasmax | gn | NaN | 20200528 |
| 437550 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | tasmin | gn | NaN | 20200528 |
| 437555 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | tas | gn | NaN | 20200528 |
| 437687 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | hurs | gn | NaN | 20200528 |
| 437792 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | pr | gn | NaN | 20200528 |
| 437906 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | pr | gn | NaN | 20200528 |
| 437907 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | huss | gn | NaN | 20200528 |
| 437918 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | hurs | gn | NaN | 20200528 |
| 437977 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | tasmin | gn | NaN | 20200528 |
| 437978 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | tasmax | gn | NaN | 20200528 |
| 437979 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | tas | gn | NaN | 20200528 |
| 438988 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | tas | gn | NaN | 20200528 |
| 439008 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | tas | gn | NaN | 20200528 |
| 439033 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | tasmax | gn | NaN | 20200528 |
| 439034 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | tasmin | gn | NaN | 20200528 |
| 439072 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | pr | gn | NaN | 20200528 |
| 439073 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | huss | gn | NaN | 20200528 |
| 439075 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | hurs | gn | NaN | 20200528 |
| 439080 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | tasmax | gn | NaN | 20200528 |
| 439227 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | hurs | gn | NaN | 20200528 |
| 439229 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | huss | gn | NaN | 20200528 |
| 439230 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | pr | gn | NaN | 20200528 |
| 439241 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | tasmin | gn | NaN | 20200528 |
# Assess unique values of ncar_models
meval = cpull.model_eval(ncar_models)
(28, 11)
activity_id : ['ScenarioMIP', 'CMIP']
institution_id : ['NCAR']
source_id : ['CESM2']
experiment_id : ['ssp370', 'historical', 'ssp245', 'ssp126', 'ssp585']
member_id : ['r11i1p1f1']
table_id : ['Amon']
variable_id : ['tasmin', 'huss', 'tas', 'hurs', 'pr', 'tasmax']
grid_label : ['gn']
version : [20200528, 20190514]
| activity_id | institution_id | source_id | experiment_id | member_id | table_id | variable_id | grid_label | zstore | dcpp_init_year | version | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 198051 | CMIP | NCAR | CESM2 | historical | r11i1p1f1 | Amon | hurs | gn | gs://cmip6/CMIP6/CMIP/NCAR/CESM2/historical/r1... | NaN | 20190514 |
| 437550 | ScenarioMIP | NCAR | CESM2 | ssp126 | r11i1p1f1 | Amon | tasmin | gn | gs://cmip6/CMIP6/ScenarioMIP/NCAR/CESM2/ssp126... | NaN | 20200528 |
| 437918 | ScenarioMIP | NCAR | CESM2 | ssp585 | r11i1p1f1 | Amon | hurs | gn | gs://cmip6/CMIP6/ScenarioMIP/NCAR/CESM2/ssp585... | NaN | 20200528 |
| 439033 | ScenarioMIP | NCAR | CESM2 | ssp370 | r11i1p1f1 | Amon | tasmax | gn | gs://cmip6/CMIP6/ScenarioMIP/NCAR/CESM2/ssp370... | NaN | 20200528 |
| 439227 | ScenarioMIP | NCAR | CESM2 | ssp245 | r11i1p1f1 | Amon | hurs | gn | gs://cmip6/CMIP6/ScenarioMIP/NCAR/CESM2/ssp245... | NaN | 20200528 |
# Use the grab_data() function to check the output of one model variable
ds = cpull.grab_data(ncar_models, 439230)
ds
<xarray.Dataset>
Dimensions: (lat: 192, nbnd: 2, lon: 288, time: 1032)
Coordinates:
* lat (lat) float64 -90.0 -89.06 -88.12 -87.17 ... 88.12 89.06 90.0
lat_bnds (lat, nbnd) float64 dask.array<chunksize=(192, 2), meta=np.ndarray>
* lon (lon) float64 0.0 1.25 2.5 3.75 5.0 ... 355.0 356.2 357.5 358.8
lon_bnds (lon, nbnd) float64 dask.array<chunksize=(288, 2), meta=np.ndarray>
* time (time) object 2015-01-15 12:00:00 ... 2100-12-15 12:00:00
time_bnds (time, nbnd) object dask.array<chunksize=(1032, 2), meta=np.ndarray>
Dimensions without coordinates: nbnd
Data variables:
pr (time, lat, lon) float32 dask.array<chunksize=(288, 192, 288), meta=np.ndarray>
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: ScenarioMIP
branch_method: standard
branch_time_in_child: 735110.0
branch_time_in_parent: 735110.0
case_id: 1731
... ...
tracking_id: hdl:21.14100/077f7679-2555-431e-a864-29011759e8e7...
variable_id: pr
variant_info: CMIP6 SSP2-4.5 experiments (2015-2100) with CAM6,...
variant_label: r11i1p1f1
netcdf_tracking_ids: hdl:21.14100/077f7679-2555-431e-a864-29011759e8e7...
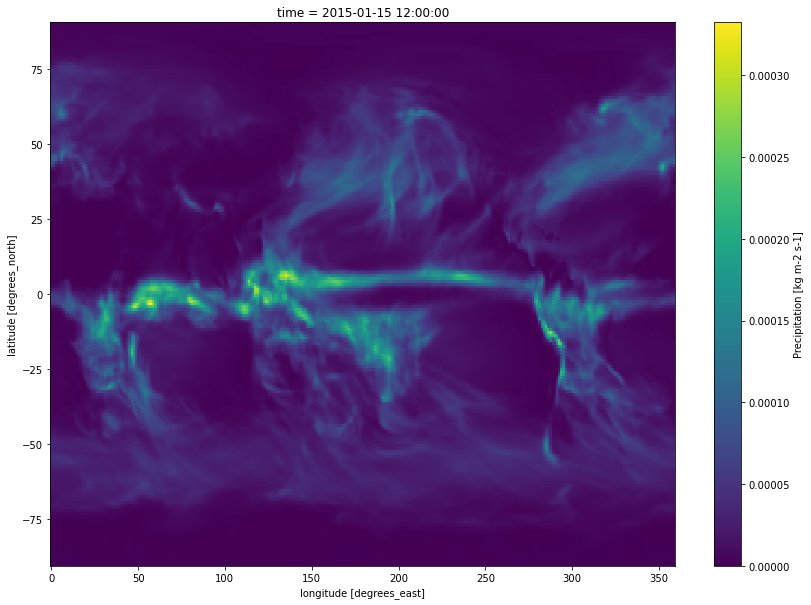
version_id: v20200528# Plot the model variable (precipitation) at one timestamp
ds.sel(time='2015-01-15')['pr'].plot(figsize=(14, 10));
# Assess the selected model variables to check for consistency in dims/coords
## All CMIP data have same coords, and all ScenarioMIP data have same coords
cpull.data_eval(ncar_models)
| activity_id | experiment_id | variable_id | lat_size | lat_min | lat_max | lon_size | lon_min | lon_max | time_size | time_min | time_max | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 198051 | CMIP | historical | hurs | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1980 | 1850-01-15 12:00:00 | 2014-12-15 12:00:00 |
| 198054 | CMIP | historical | huss | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1980 | 1850-01-15 12:00:00 | 2014-12-15 12:00:00 |
| 198057 | CMIP | historical | pr | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1980 | 1850-01-15 12:00:00 | 2014-12-15 12:00:00 |
| 198172 | CMIP | historical | tas | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1980 | 1850-01-15 12:00:00 | 2014-12-15 12:00:00 |
| 437687 | ScenarioMIP | ssp126 | hurs | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437199 | ScenarioMIP | ssp126 | huss | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437792 | ScenarioMIP | ssp126 | pr | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437555 | ScenarioMIP | ssp126 | tas | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437549 | ScenarioMIP | ssp126 | tasmax | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437550 | ScenarioMIP | ssp126 | tasmin | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439227 | ScenarioMIP | ssp245 | hurs | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439229 | ScenarioMIP | ssp245 | huss | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439230 | ScenarioMIP | ssp245 | pr | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 438988 | ScenarioMIP | ssp245 | tas | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439080 | ScenarioMIP | ssp245 | tasmax | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439241 | ScenarioMIP | ssp245 | tasmin | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439075 | ScenarioMIP | ssp370 | hurs | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439073 | ScenarioMIP | ssp370 | huss | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439072 | ScenarioMIP | ssp370 | pr | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439008 | ScenarioMIP | ssp370 | tas | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439033 | ScenarioMIP | ssp370 | tasmax | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 439034 | ScenarioMIP | ssp370 | tasmin | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437918 | ScenarioMIP | ssp585 | hurs | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437907 | ScenarioMIP | ssp585 | huss | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437906 | ScenarioMIP | ssp585 | pr | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437979 | ScenarioMIP | ssp585 | tas | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437978 | ScenarioMIP | ssp585 | tasmax | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
| 437977 | ScenarioMIP | ssp585 | tasmin | 192 | -90.0 | 90.0 | 288 | 0.0 | 358.75 | 1032 | 2015-01-15 12:00:00 | 2100-12-15 12:00:00 |
# Check how many variables each model has
## The 2 extra in ScenarioMIP are for 'tasmin' and 'tasmax'
model_groups = pd.DataFrame({
'# variables': ncar_models.groupby(['activity_id', 'experiment_id']).size()
})
model_groups
| # variables | ||
|---|---|---|
| activity_id | experiment_id | |
| CMIP | historical | 4 |
| ScenarioMIP | ssp126 | 6 |
| ssp245 | 6 | |
| ssp370 | 6 | |
| ssp585 | 6 |
# Read in the selected model data!
if os.path.exists('../data/historical.zarr'):
print('Data already downloaded!')
else:
cpull.model_pull()
Completed downloading ('CMIP', 'historical')!
Completed downloading ('ScenarioMIP', 'ssp126')!
Completed downloading ('ScenarioMIP', 'ssp245')!
Completed downloading ('ScenarioMIP', 'ssp370')!
Completed downloading ('ScenarioMIP', 'ssp585')!
